Research
Theme 1
Elucidating the working principles of the genome
 The genome plays a central role in life, enabling numerous biological functions such as differentiation, development, and immune and disease responses. However, our understanding of its underlying mechanisms remains incomplete, and predicting how drugs can control genome function is still a major challenge.
The genome plays a central role in life, enabling numerous biological functions such as differentiation, development, and immune and disease responses. However, our understanding of its underlying mechanisms remains incomplete, and predicting how drugs can control genome function is still a major challenge.
Our laboratory investigates the molecular-level architecture of the genome to elucidate the physical and chemical foundations by which genes encoded in the genome are expressed. Building on this knowledge, we aim to develop methodologies for logically controlling these processes through pharmacological interventions.
The Hi-CO method
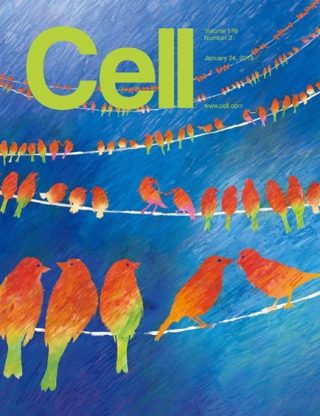
We recently developed the Hi-CO method to clarify the 3D molecular structure of the genome (Ohno et al., Cell, 2019). This method combines measurements of genomic structural information with next-generation genome sequencing and the simulation of molecular dynamics using a supercomputer to analyze the 3D arrangement and orientation of every nucleosome over the entire genome. It was already known that the genome wraps around histone octamer proteins at every 160 to 200 base pairs to form a unitary structure called the nucleosome. The Hi-CO method has expanded on this knowledge and revealed that the nucleosome folding structure is significantly altered at every genomic locus according to gene regulation status. This work has been prominently featured on the cover of Cell. For more details, please refer to https://www.riken.jp/en/news_pubs/research_news/rr/20190705_FY20190014/.
Derivation of 3D nucleosome folding structure by simulated annealing molecular dynamics simulation (Ohno et al., Cell, 2019)
The BHi-Cect method
The genome is comprised of macromolecules formed by very long series of nucleosomes with a hierarchically organized structure within the nucleus of the cell. The genome is known to form domain substructures at the multiple gene locus level, and to form loop substructures around transcription initiation sites. This complex structure exists in two structural states, a highly condensed state and a dispersed state, which are involved in gene inactivation and activation, respectively. In our laboratory, we have developed an algorithm, the BHi-Cect method, that identifies characteristic structural elements from the genome using machine learning (Kumar et al., Nucleic Acids Research, 2020). Using this method, we have discovered novel structural elements (“enclaves”) and demonstrated that they are tightly related to gene activity. For more information, please see https://www.riken.jp/en/news_pubs/research_news/rr/20200501_1/index.html.
Theme 2
Understanding the constitutional principles of cellular systems
 Cells, the fundamental units of life, are composed of an enormous diversity of molecules. Among them, the ~20,000 proteins encoded by the genome play a crucial role in shaping the functional states of cells. Our laboratory is developing methods to comprehensively and quantitatively reveal the molecular composition of individual cells. By doing so, we aim to elucidate the mechanisms by which cells manifest diverse functions, while also creating new diagnostic approaches that sensitively detect disease signatures arising from errors in these mechanisms.
Cells, the fundamental units of life, are composed of an enormous diversity of molecules. Among them, the ~20,000 proteins encoded by the genome play a crucial role in shaping the functional states of cells. Our laboratory is developing methods to comprehensively and quantitatively reveal the molecular composition of individual cells. By doing so, we aim to elucidate the mechanisms by which cells manifest diverse functions, while also creating new diagnostic approaches that sensitively detect disease signatures arising from errors in these mechanisms.
Imaging single-molecule gene expression in single cells
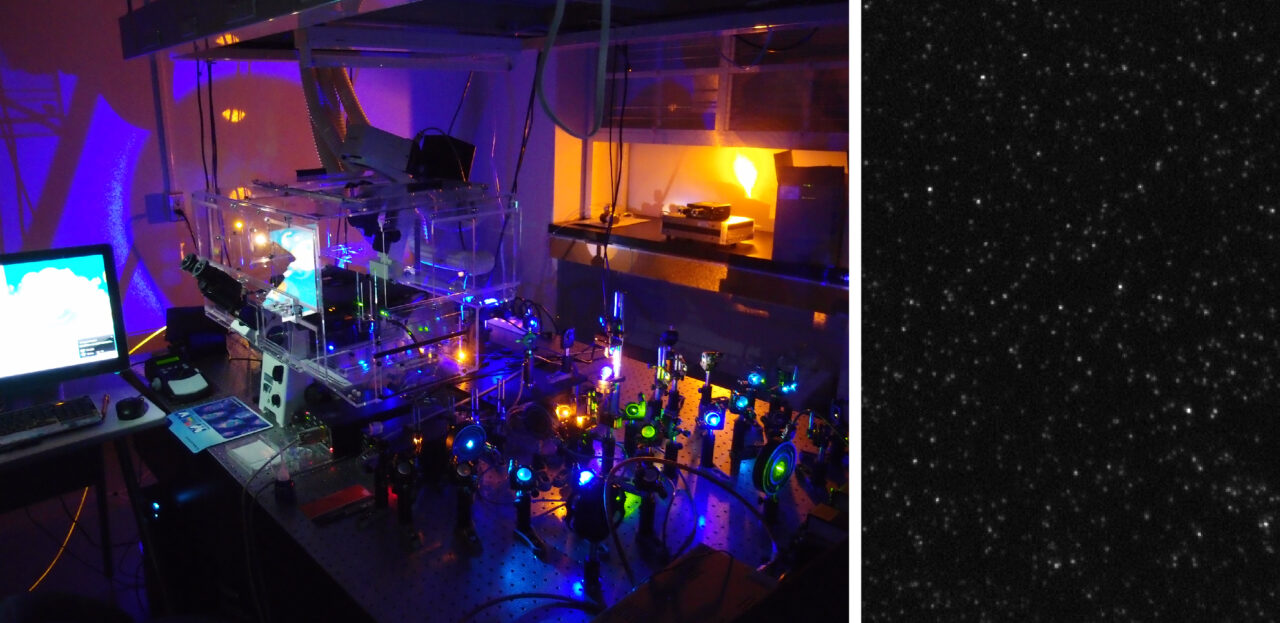
It is known that every single cell, even if it is genetically identical to other cells, exists in its own unique state. These unique states are achieved by the expression of different quantities of proteins. To illuminate the logic of intracellular gene regulation, we must be able to examine single-cell dynamics with high accuracy. Our laboratory developed a method to analyze single cell gene expression dynamics with single-molecule resolution for a large number of genes (Taniguchi et al., Science, 2010). Our technique involves a high-throughput platform that combines automated single-molecule fluorescence microscopy and a cell library of chromosomal fluorescent protein fusion. Using this technique, we discovered that there is a general lack of correlation between the quantity of mRNA and the quantity of proteins in a single cell.
PISA microscopy
Single-
Although mass spectrometry is a standard method for proteome analysis, it presents issues, such as low sensitivity, low throughput, and high equipment cost that make it less than ideal for the diagnosis of disease. Our laboratory seeks methodological solutions to these issues. For example, we are developing the use of single molecule microscopy to improve sensitivity (Leclerc et al., Bioconj. Chem., 2018; Taniguchi, Leclerc: Patent Application JP2017-177070; Taniguchi, Ohno: PCT/JP2018/048329). We expect that this technique will eventually enable the high-throughput analysis of small amounts of samples (even single cells) via an automated system.
Theme 3
Predicting cellular responses at the molecular level with AI
 To accurately capture and flexibly control the state of life, it is essential to integrate the vast molecular information obtained from the cells. Our laboratory employs cutting-edge AI technologies and mathematical theories to construct detailed models of cellular systems. This project is being conducted as a JST CREST program led by Professor Taniguchi.
To accurately capture and flexibly control the state of life, it is essential to integrate the vast molecular information obtained from the cells. Our laboratory employs cutting-edge AI technologies and mathematical theories to construct detailed models of cellular systems. This project is being conducted as a JST CREST program led by Professor Taniguchi.